Martin Karplus, Michael Levitt, and Arieh Warshel won the 2013 Nobel Prize For Chemistry today "for the development of multiscale models for complex chemical systems."
The Royal Swedish Academy of Sciences said the three scientists' research in the 1970s has helped scientists develop programs that unveil chemical processes. "The work of Karplus, Levitt and Warshel is ground-breaking in that they managed to make Newton's classical physics work side-by-side with the fundamentally different quantum physics," the academy said. "Previously, chemists had to choose to use either/or." Together with a few earlier Nobel Prizes in quantum chemistry, this award consecrates the field of computational chemistry.
 Incidentally, Martin Karplus is my postdoc co-adviser Georgios Archontis's thesis adviser at Harvard. Georgios is one of the earlier contributors to CHARMM, a widely-used package of computational chemistry. CHARMM was the computational tool that I used when working with Georgios almost 15 years ago. In collaboration with Martin, Georgios and I were studying glycogen phosphorylase inhibitors based on a free energy perturbation analysis using CHARMM. In another project with Spyros Skourtis, I wrote a multi-scale simulation program that couples molecular dynamics and quantum dynamics to study electron transfer in proteins and DNA molecules (i.e., use Newton's Equation of Motion to predict the trajectories of atoms, construct the Hamiltonian time series, and solve the time-dependent Schrodinger equation using the Hamiltonian series as the input).
Incidentally, Martin Karplus is my postdoc co-adviser Georgios Archontis's thesis adviser at Harvard. Georgios is one of the earlier contributors to CHARMM, a widely-used package of computational chemistry. CHARMM was the computational tool that I used when working with Georgios almost 15 years ago. In collaboration with Martin, Georgios and I were studying glycogen phosphorylase inhibitors based on a free energy perturbation analysis using CHARMM. In another project with Spyros Skourtis, I wrote a multi-scale simulation program that couples molecular dynamics and quantum dynamics to study electron transfer in proteins and DNA molecules (i.e., use Newton's Equation of Motion to predict the trajectories of atoms, construct the Hamiltonian time series, and solve the time-dependent Schrodinger equation using the Hamiltonian series as the input).
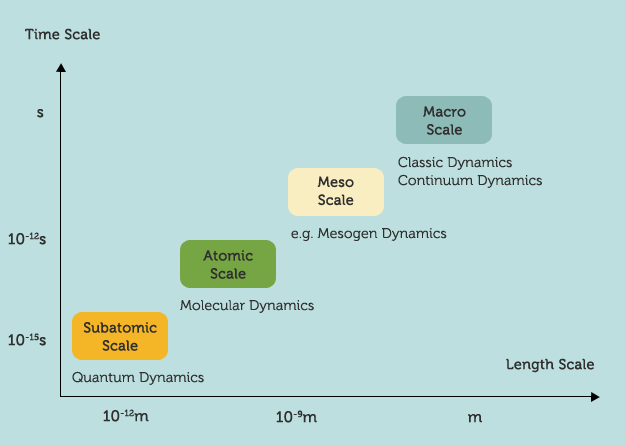
We are thrilled by this news because much of the computational kernels of our Molecular Workbench software was actually inspired by CHARMM. The Molecular Workbench also advocates a multiscale philosophy and pedagogical approach, but for linking concepts at different scales with simulations in order to help students connect the dots and build more unified pictures about science (see the image above).
We are glad to be part of the "Karplus genealogy tree," as Georgios put it when replying my congratulatory email. We hope that through our grassroots work in education, the power of molecular simulation from the top of the scientific research pyramid will enlighten millions of students and ignite their interest and curiosity in science.
The Royal Swedish Academy of Sciences said the three scientists' research in the 1970s has helped scientists develop programs that unveil chemical processes. "The work of Karplus, Levitt and Warshel is ground-breaking in that they managed to make Newton's classical physics work side-by-side with the fundamentally different quantum physics," the academy said. "Previously, chemists had to choose to use either/or." Together with a few earlier Nobel Prizes in quantum chemistry, this award consecrates the field of computational chemistry.
 Incidentally, Martin Karplus is my postdoc co-adviser Georgios Archontis's thesis adviser at Harvard. Georgios is one of the earlier contributors to CHARMM, a widely-used package of computational chemistry. CHARMM was the computational tool that I used when working with Georgios almost 15 years ago. In collaboration with Martin, Georgios and I were studying glycogen phosphorylase inhibitors based on a free energy perturbation analysis using CHARMM. In another project with Spyros Skourtis, I wrote a multi-scale simulation program that couples molecular dynamics and quantum dynamics to study electron transfer in proteins and DNA molecules (i.e., use Newton's Equation of Motion to predict the trajectories of atoms, construct the Hamiltonian time series, and solve the time-dependent Schrodinger equation using the Hamiltonian series as the input).
Incidentally, Martin Karplus is my postdoc co-adviser Georgios Archontis's thesis adviser at Harvard. Georgios is one of the earlier contributors to CHARMM, a widely-used package of computational chemistry. CHARMM was the computational tool that I used when working with Georgios almost 15 years ago. In collaboration with Martin, Georgios and I were studying glycogen phosphorylase inhibitors based on a free energy perturbation analysis using CHARMM. In another project with Spyros Skourtis, I wrote a multi-scale simulation program that couples molecular dynamics and quantum dynamics to study electron transfer in proteins and DNA molecules (i.e., use Newton's Equation of Motion to predict the trajectories of atoms, construct the Hamiltonian time series, and solve the time-dependent Schrodinger equation using the Hamiltonian series as the input). We are thrilled by this news because much of the computational kernels of our Molecular Workbench software was actually inspired by CHARMM. The Molecular Workbench also advocates a multiscale philosophy and pedagogical approach, but for linking concepts at different scales with simulations in order to help students connect the dots and build more unified pictures about science (see the image above).
We are glad to be part of the "Karplus genealogy tree," as Georgios put it when replying my congratulatory email. We hope that through our grassroots work in education, the power of molecular simulation from the top of the scientific research pyramid will enlighten millions of students and ignite their interest and curiosity in science.

